Bioinformatic; Learn Bulk RNA-Seq Data Analysis From Scratch
Muhammad Dujana
4:40:29
Description
Best Bioinformatics course for Students, Academia and Industry Professionals to learn RNA-Seq Data Analysis From Zero
What You'll Learn?
- You will be able to understand basic molecular biology; Central Dogma
- You will be able to Understand RNA-Seq Experimentation
- You will be able to analyze FASTQ files In Linux Environment
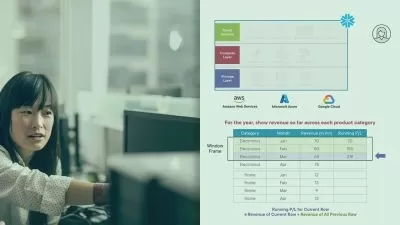
- You will be able to understand different file formats like SAM, BAM, FASTQ, GTF, etc
- You will be able to Use R and R-Studio
- You will able to perform Differential analysis of Genes using DESeq2 Package
- You will able to generate Different type of visualization to present your Data like PCA, MA, HeatMap and Volcano Plots
- You will be able to perform GO and Pathways Analysis
Who is this for?
What You Need to Know?
More details
DescriptionWelcome to our third course "Learn Bulk RNA-Seq Data Analysis From Scratch," a comprehensive online course designed to equip you with the skills and knowledge needed to harness the power of RNA-Seq data analysis. In this course, we delve into the captivating world of genomics and bioinformatics, empowering you to explore the intricacies of gene expression and unravel the hidden mysteries within the transcriptome.
With the advent of high-throughput sequencing technologies, RNA-Seq has revolutionized the field of molecular biology, allowing us to decipher the intricate dance of gene expression in ways never before possible. This course serves as your gateway to understanding and interpreting the wealth of information contained within RNA-Seq data, transforming it into valuable insights and meaningful discoveries.
Bioinformatics, the multidisciplinary field at the intersection of biology and computer science, plays a pivotal role in deciphering complex biological systems. In this course, we emphasize the importance of bioinformatics methodologies and tools, which form the foundation of modern genomics research. By mastering these techniques, you will gain a competitive edge in the rapidly evolving field of life sciences.
Course Highlights:
Comprehensive Training: From raw FASTQ files to in-depth analysis, this course provides a step-by-step guide to RNA-Seq data analysis, covering the entire workflow with clarity and precision.
Linux and R-Studio: Get hands-on experience with two essential tools in bioinformatics. Learn to navigate the Linux command line environment and utilize R-Studio for data processing, visualization, and statistical analysis.
Theory and Practice: We strike a perfect balance between theoretical concepts and practical application. Understand the underlying principles of RNA-Seq analysis while honing your skills through hands-on exercises and real-world examples.
Cutting-edge Techniques: Stay at the forefront of genomics research by exploring the latest advancements in RNA-Seq analysis techniques, such as differential gene expression analysis, functional enrichment analysis, and pathway analysis.
Expert Guidance: Benefit from the expertise of experienced instructors who have a deep understanding of both bioinformatics and molecular biology. Their guidance and insights will ensure a rewarding learning experience.
Interactive Learning: Engage in interactive assignments, and discussions to reinforce your understanding and interact with a vibrant community of fellow learners, fostering knowledge exchange.
Embark on this transformative journey into the world of RNA-Seq analysis and bioinformatics. Unleash the power of genomics to uncover hidden biological insights and make significant contributions to scientific research. Enroll in "Bioinformatics: Learn Bulk RNA-Seq Data Analysis From Scratch" today and equip yourself with the essential skills needed to excel in the dynamic field of bioinformatics. We assure you that all of the tools that will be used in this course will be Freely available and closely related to the course material. For most of them you do not need to sign up.
Who this course is for:
- Molecular Biologist
- Biological Data Analyst
- Bioinformatics
Welcome to our third course "Learn Bulk RNA-Seq Data Analysis From Scratch," a comprehensive online course designed to equip you with the skills and knowledge needed to harness the power of RNA-Seq data analysis. In this course, we delve into the captivating world of genomics and bioinformatics, empowering you to explore the intricacies of gene expression and unravel the hidden mysteries within the transcriptome.
With the advent of high-throughput sequencing technologies, RNA-Seq has revolutionized the field of molecular biology, allowing us to decipher the intricate dance of gene expression in ways never before possible. This course serves as your gateway to understanding and interpreting the wealth of information contained within RNA-Seq data, transforming it into valuable insights and meaningful discoveries.
Bioinformatics, the multidisciplinary field at the intersection of biology and computer science, plays a pivotal role in deciphering complex biological systems. In this course, we emphasize the importance of bioinformatics methodologies and tools, which form the foundation of modern genomics research. By mastering these techniques, you will gain a competitive edge in the rapidly evolving field of life sciences.
Course Highlights:
Comprehensive Training: From raw FASTQ files to in-depth analysis, this course provides a step-by-step guide to RNA-Seq data analysis, covering the entire workflow with clarity and precision.
Linux and R-Studio: Get hands-on experience with two essential tools in bioinformatics. Learn to navigate the Linux command line environment and utilize R-Studio for data processing, visualization, and statistical analysis.
Theory and Practice: We strike a perfect balance between theoretical concepts and practical application. Understand the underlying principles of RNA-Seq analysis while honing your skills through hands-on exercises and real-world examples.
Cutting-edge Techniques: Stay at the forefront of genomics research by exploring the latest advancements in RNA-Seq analysis techniques, such as differential gene expression analysis, functional enrichment analysis, and pathway analysis.
Expert Guidance: Benefit from the expertise of experienced instructors who have a deep understanding of both bioinformatics and molecular biology. Their guidance and insights will ensure a rewarding learning experience.
Interactive Learning: Engage in interactive assignments, and discussions to reinforce your understanding and interact with a vibrant community of fellow learners, fostering knowledge exchange.
Embark on this transformative journey into the world of RNA-Seq analysis and bioinformatics. Unleash the power of genomics to uncover hidden biological insights and make significant contributions to scientific research. Enroll in "Bioinformatics: Learn Bulk RNA-Seq Data Analysis From Scratch" today and equip yourself with the essential skills needed to excel in the dynamic field of bioinformatics. We assure you that all of the tools that will be used in this course will be Freely available and closely related to the course material. For most of them you do not need to sign up.
Who this course is for:
- Molecular Biologist
- Biological Data Analyst
- Bioinformatics
User Reviews
Rating
Muhammad Dujana
Instructor's Courses
Udemy
View courses Udemy- language english
- Training sessions 61
- duration 4:40:29
- Release Date 2023/08/19